Introduction
In the previous section of this series, we went through the setup, customization of Tools and layout, and the first run of the Chimera application. We also employed simple scripts and introduced keyboard shortcuts.
Part-3 features details on using the navigation Menu options, command functions, and accelerator keys. Both simple and more advanced steps will be demonstrated on the Collapsin Response Mediator Protein 2 (CRMP2) - protein of rising pharmaceutical relevance.
Using the Chimera
Since many detailed tutorials and websites demonstrate both basic and advanced moves in Chimera, we will focus on steps and commands that were not entirely intuitive to me or were not sufficiently documented. Chimera users discussion mailing list is an exceptional source of information, tips, and tweaks. The list topics are best searched using Google.
First, the list of excellent tutorials, descriptions, and official Chimera pages:
Recommended
- 3D Structure Visualization with Chimera - pdf - start here
- Computer graphics lab, UCSF tutorials
- Chimera User's Guide
- QUICK REFERENCE GUIDE
- UCSF Chimera - I - Introduction
- UCSF Chimera - Getting Started
- RBVI Molecular Visualization - pdf
Figure 1 shows the essential Menu items and places where commands can be issued. The green dotted rectangles outline menu items that are used at the beginning of each session. The menu expands in the solid blue rectangle are used very frequently for selections and rendering. While the Menu items are a powerful feature, the more one gets familiar with the Chimera's "logic", the more one appreciates the use of Command line and Keyboard accelerators.
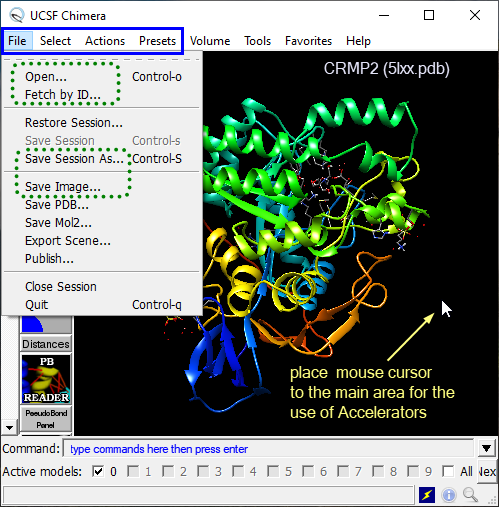
Figure 1. Chimera's workspace highlighting the main places for navigation and visualization (Menu expands, Command line, Keyboard accelerators).
Let's start with steps, such as Opening/Loading pdb file, selecting one-two atoms, the whole amino acid residue or one of the chains. Next, we will remove water molecules, solvents, counter ions, and ligand in the active site.
Most significantly, the trafficking and activity of Nav1.7 are attenuated by SUMOylation of K374 that is located in the CRMP2 monomer at the loop flanked by helix residues 360-371 and 376-392. Considering the genetic validation of Nav1.7 as a pain target, small molecule inhibitors of K374 SUMOylation by Ubc9 E2 SUMO-conjugating enzyme may offer a novel approach to non-opioid pain therapeutics. Indeed, indirect and a dose-dependent modulation of conductivity and trafficking of sodium channel Nav1.7 has been shown to reverse neuropatic pain in rats.
Drug discovery program targeting such interaction is currently progressing at the Regulonix, and indirect small molecule modulators of Nav1.7 are being developed by the team headed by Dr. Rajesh Khanna.
Let's start with selections. I chose a not so trivial example, which includes multiple protein chains and residue conformers (quite a typical situation). Since we have already defined alias style in Part 2, we will use it from the Command line to improve the visual appearance of CRMP2 monomer.
Preparation:
- start the Chimera application
- using the Menu items or Command line (Fig. 1), execute the first five rows in the Table 1 below
- Next, type
focus :374into the Command line, press enter followed bydisplay :374with enter. This will show two side-chains of Lys 374 (example) and center the view at them. Type~display HCto hide nonpolar hydrogens. - type
~display :374@*.Bto hide the other (calculated) conformer (B) of Lys 374 - optionally, point the mouse cursor somewhere to the blank workspace and press
bson the keyboard (the side-chain should be rendered asball and stick) - use left mouse button to orient residues of Lys 374 to resemble images in Figure 2
- use the mouse wheel (forward/backward) OR the right mouse button to zoom in/out
- use the mouse wheel (press it) to move the molecule in X-Y direction
- type
clip offreset the plane clipping toauto(or move the clipping plane by mouse by starting the Clipping plane panel - icons on the left)
The corresponding Chimera session file that runs through the Preparation above can be downloaded here. To load the session file, unzip the file and use the File → Restore session menu item in Figure 1.
Selections:
- to select one atom, point the mouse cursor at it and press Ctrl + left mouse button (Fig. 2, left)
- to select (and add) another atom, press Ctrl + Shift keys and left-mouse click it (Fig. 2, middle)
- to expand selection to the whole residue, press the
up keyon your keyboard (Fig. 2, right) - to deselect one or more selected objects, point the mouse cursor to the blank workspace, press Ctrl key and left-click by mouse, or press once the
upand several times thedownkey on the keyboard, or typecs(clear selection)
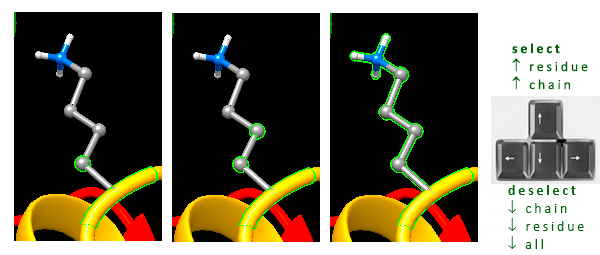
Fig. 2. Simple atom and residue selections using Ctrl and Shift keys with the left mouse click.
The sequence of actions in the table below follows a typical workflow of protein visualization, including the file opening, getting the protein monomer, coloring by secondary structure, removing solvent, and salts to saving the session and making an image.
Run the options in Table 1, either from the Menu, Command line, or by Accelerator keys to get identical or similar results. The options can be combined.
Table 1.
| Action | Menu Item | Command | Accelerator keys (type `ad` to list) |
|---|---|---|---|
| OPEN FILE | File → Open... (5lxx.pdb) or File → Fetch by ID... (from the Web, 5lxx) | open /path/to/5lxx.pdb | op or ff (fetch) |
| DELETE chain B | Select → Chain → B; Action → Atoms/Bonds → delete | del :.B-F (delete all but A chains) | - |
| style it | Actions → Focus; Tools → Depiction → Color Secondary Structure; Actions → Atoms/Bonds → wire; Actions → Color → by element; Select → Structure → protein; Action → Atoms/Bonds → hide |
style OR run style.py ~display protein (hide side chains) |
fo (focus on center of protein) c2 or rc (color by secondary structures) wr (wireframe representation) ce (color by atom-type) bb (show backbone only) |
| orient | use left mouse + middle wheel (press) to orient | focus turn x 90 turn z 90 |
x9, z9 (rotate by 90 deg about x, z axes) - run one key pair after another (no commas) |
| clean it | Select → Residue → SO4 Action → Atoms/Bonds → delete |
del :SO4 del solvent |
- |
| save it | File → Save Image (as .png, rendering: Chimera, supersample 3x3) File → Save Session as |
copy file ~/Desktop/test.png png dpi 300 supersample 3 (change the path in blue) save (save session as) |
si (save image) ss (save session as) |
There are three ways to select objects [atom(s), residue(s), chain(s), model(s)] in Chimera.
- A. by left mouse click with Ctrl key pressed simultaneously; and adding to this selection by holding Shift + Ctrl key followed by the left mouse click
- B. selecting from the menu expand `Select` at the top of the GUI
- C. using Command line syntax
Selecting by Commands
Commands are a powerful and compelling option in structure/model manipulation. Examples of the most typical selectors follow:
type select (or sel) into the Command line followed by space, like so:
sel #0 selects object 0 (typically the protein; all atoms)
sel #0.1 selects submodel 0.1 (usually after splitting the model with command `split`)
sel :374 selects amino acid residue number 374
sel :cys selects all Cys residues
sel :SO4 selects sulfate anions
sel solvent select solvent molecules (typically water)
sel :.A selects chain A
sel :374 & @NZ select Lys374 and its nitrogen atom (hover mouse cursor over nitrogen to identify its ID; if charged in the pdb file, use :374 & N3+)
sel :374@*.A select Lys374 and all atoms (*) in conformer A
sel :374-376,378 select residues 374 to 376 and residue 378 (hover mouse cursor over amino acid to identify its ID)
sel :ile/isHelix select all Ile residues in helices (capital H is required)
sel :ile,tyr/isStrand select all Ile and Tyr residues in beta-strands
To reverse the function (command), use ~ followed by the command (no space), like so:
~sel :374 # deselect Lys374
~display protein # hide amino acids in the protein
~disp element.H # hide all hydrogens
~rlabel # remove residue labels
~ribbon # hide the ribbon representation of protein
Examples of other commands that I use frequently are shown below with descriptive comments:
# Show only polar hydrogens (2-step procedure)
display element.H # show all hydrogens
~disp HC # hide non-polar hydrogens
# Render surface of the ligand cavity
# continue after executing the top five rows in Table 1
sel :BTB # select ligand - hover mouse cursor over the ligand to get its ID-name
represent bs sel # represent the selection (ligand) as ball-and-stick
~disp HC
zone sel 5 # select atoms around the selected ligand BTB (sel) within 5 Å
sel up # select whole amino acid residues
disp sel # display selected objects
~disp solvent
surface sel # generate cavity surface - including selected residues
# color the surface by hydrophobicity of residues - magenta polar, tan hydrophobic
rangecolor kdHydrophobicity min medium purple 0 white max tan sel
~ribbon # hide ribbon rendering
rlabel sel # label selected residues
~sel # deselect all
The semicolon separator (;) concatenates the sequence of commands. To execute functions in one line, copy/paste the command below.
# DELETE ALL chains, keep only the first one:
split; sel#0.1; sel invert; del sel; style; ~disp protein
# Rename sub-model 0.1 to model #0 and name it 'prot':
combine #0.1 model 0 name prot; close #0.1
Keyboard Accelerator
Several keyboard shortcuts were already described in Table 1. To activate the keyboard shortcuts, type ac followed by enter in the Command line. Recall that the startup script midasrc (Part 2) made the keys available.
Access the complete list of Chimera keyboard shortcuts here. Alternatively, to display a list of the keys, type ad.
The keys that I found most handy are listed next.
c2 # color by secondary structure
wb # set white background; `bk` to get back to black background
wr # wire representation (also applies to selected object)
bs # ball-and-stick representation (also applies to selected object)
hs # hide or show surface; `sf` show surface
cs # clear selection
is # invert selection (for selected Model)
iS # invert selection (all Models)
rl # open reply log (for important messages)
ce # color by element
fo # focus (also applies to selected object)
ct # show chain trace (ribbon) only (~disp protein)
c5 # select and show 5 Å contacts between selected and unselected atoms
zd # show zone dialog
mC # place marker at center of selected atoms (mark center of phenyl ring)
Next
After the introduction of Menu items, Command line functions, key shortcuts (accelerators), and after a few practical exercises, the next blog article will explore the use of custom scripts that make visualizations in UCSF Chimera faster and reproducible.
Bibliography
Joel M. Brittain, Andrew D. Piekarz, Yuying Wang, Takako Kondo, Theodore R. Cummins, and Rajesh Khanna. An atypical role for collapsin response mediator protein 2 (CRMP-2) in neurotransmitter release via interaction with presynaptic voltage-gated calcium channels. J. Biol. Chem., 284(45):31375–31390, November 2009. doi:10.1074/jbc.M109.009951. ↩
Xian Xuan Chi, Brian S. Schmutzler, Joel M. Brittain, Yuying Wang, Cynthia M. Hingtgen, Grant D. Nicol, and Rajesh Khanna. Regulation of N-type voltage-gated calcium channels (Cav2.2) and transmitter release by collapsin response mediator protein-2 (CRMP-2) in sensory neurons. J. Cell. Sci., 122(Pt 23):4351–4362, December 2009. doi:10.1242/jcs.053280. ↩
Erik T. Dustrude, Sarah M. Wilson, Weina Ju, Yucheng Xiao, and Rajesh Khanna. CRMP2 protein SUMOylation modulates NaV1.7 channel trafficking. J. Biol. Chem., 288(34):24316–24331, August 2013. doi:10.1074/jbc.M113.474924. ↩
Erik Thomas Dustrude, Samantha Perez-Miller, Liberty François-Moutal, Aubin Moutal, May Khanna, and Rajesh Khanna. A single structurally conserved SUMOylation site in CRMP2 controls NaV1.7 function. Channels (Austin), 11(4):316–328, July 2017. doi:10.1080/19336950.2017.1299838. ↩
Rajesh Khanna, Sarah M. Wilson, Joel M. Brittain, Jill Weimer, Rukhsana Sultana, Allan Butterfield, and Kenneth Hensley. Opening Pandora's jar: a primer on the putative roles of CRMP2 in a panoply of neurodegenerative, sensory and motor neuron, and central disorders. Future Neurol, 7(6):749–771, November 2012. doi:10.2217/FNL.12.68. ↩
Matti Myllykoski, Anne Baumann, Kenneth Hensley, and Petri Kursula. Collapsin response mediator protein 2: high-resolution crystal structure sheds light on small-molecule binding, post-translational modifications, and conformational flexibility. Amino Acids, 49(4):747–759, April 2017. doi:10.1007/s00726-016-2376-z. ↩
Liberty François-Moutal, Erik T. Dustrude, Yue Wang, Tatiana Brustovetsky, Angie Dorame, Weina Ju, Aubin Moutal, Samantha Perez-Miller, Nickolay Brustovetsky, Vijay Gokhale, May Khanna, and Rajesh Khanna. Inhibition of the Ubc9 E2 SUMO-conjugating enzyme-CRMP2 interaction decreases NaV1.7 currents and reverses experimental neuropathic pain. Pain, 159(10):2115–2127, October 2018. doi:10.1097/j.pain.0000000000001294. ↩
Liberty François-Moutal, David Donald Scott, Samantha Perez-Miller, Vijay Gokhale, May Khanna, and Rajesh Khanna. Chemical shift perturbation mapping of the Ubc9-CRMP2 interface identifies a pocket in CRMP2 amenable for allosteric modulation of Nav1.7 channels. Channels (Austin), 12(1):219–227, 2018. doi:10.1080/19336950.2018.1491244. ↩
Comments
comments powered by Disqus